Introduction
ODG is the Omics Database Generator.
Please see the article, ODG: Omics Database Generator for a more information. You may also download the software from GitHub.
If you are interested in using ODG at this time please contact me personally, I am happy to assist you.
Screenshots
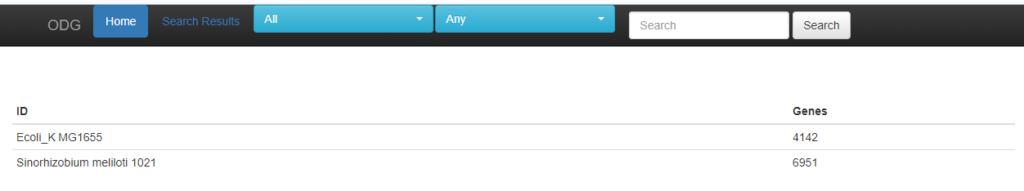
The start page of ODG lists the number of genes identified for each species. This is a good guide on whether the database has been properly generated.
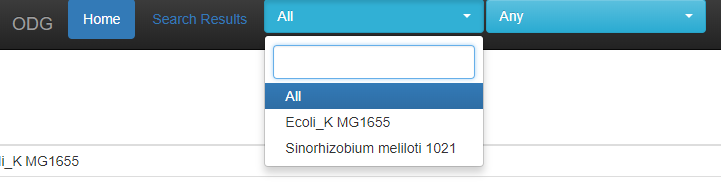
The available species are available as a drop down to query, or you may query all species in the database. A text-box autocomplete search is available as well if you have many species or accessions.
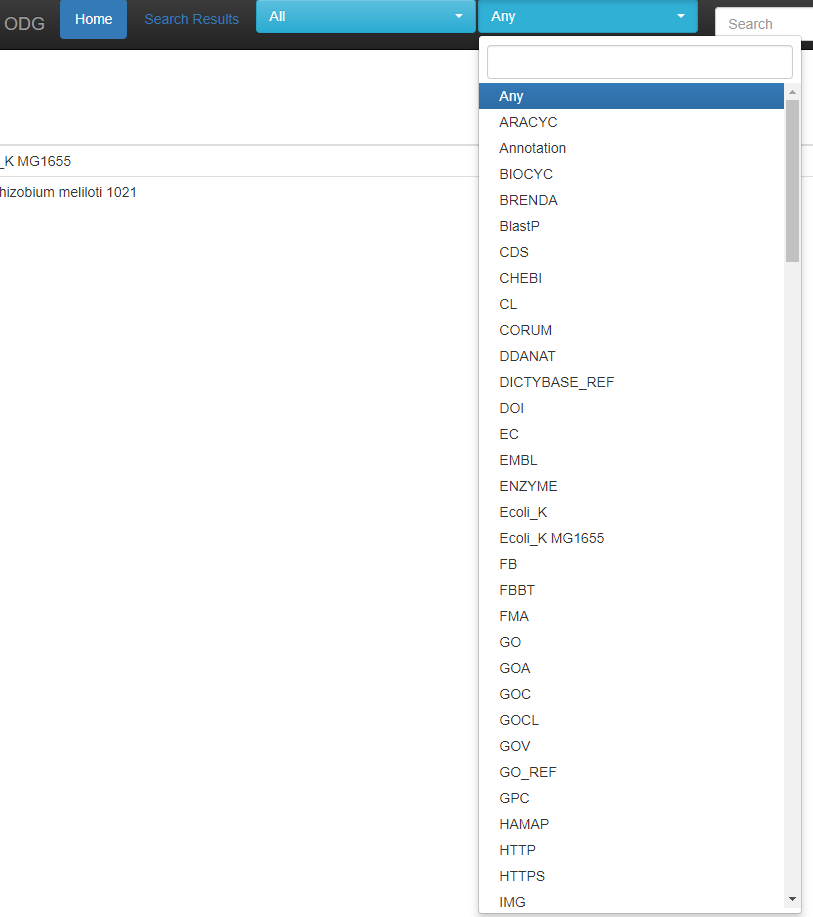
For whichever species / accession you have chosen, you may choose various labels to search on, or you may search on Any.
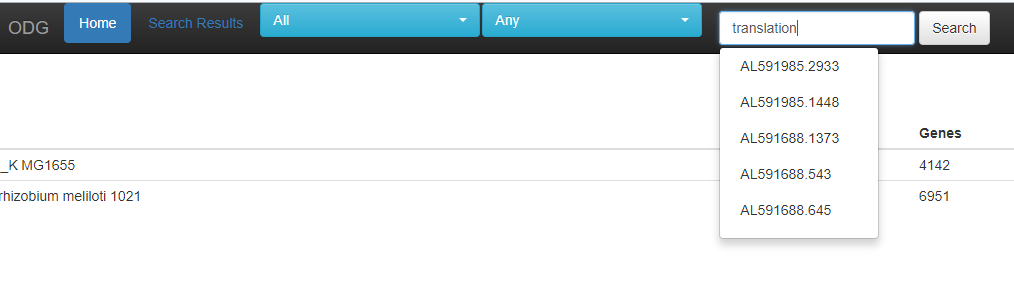
This list is updated for each species you choose, so E. coli has less options to choose from than “All” as above.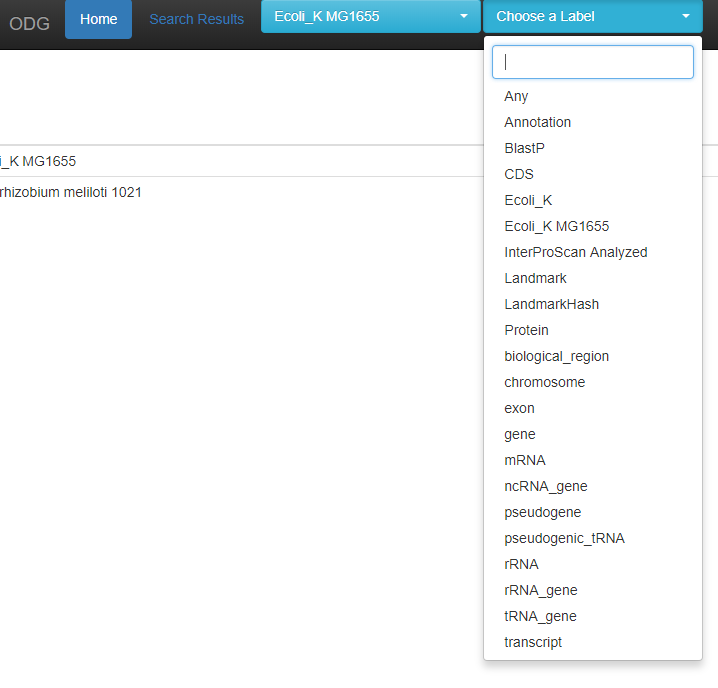 When entering a search, the first 5 options are given as suggestions for the search.
When entering a search, the first 5 options are given as suggestions for the search.
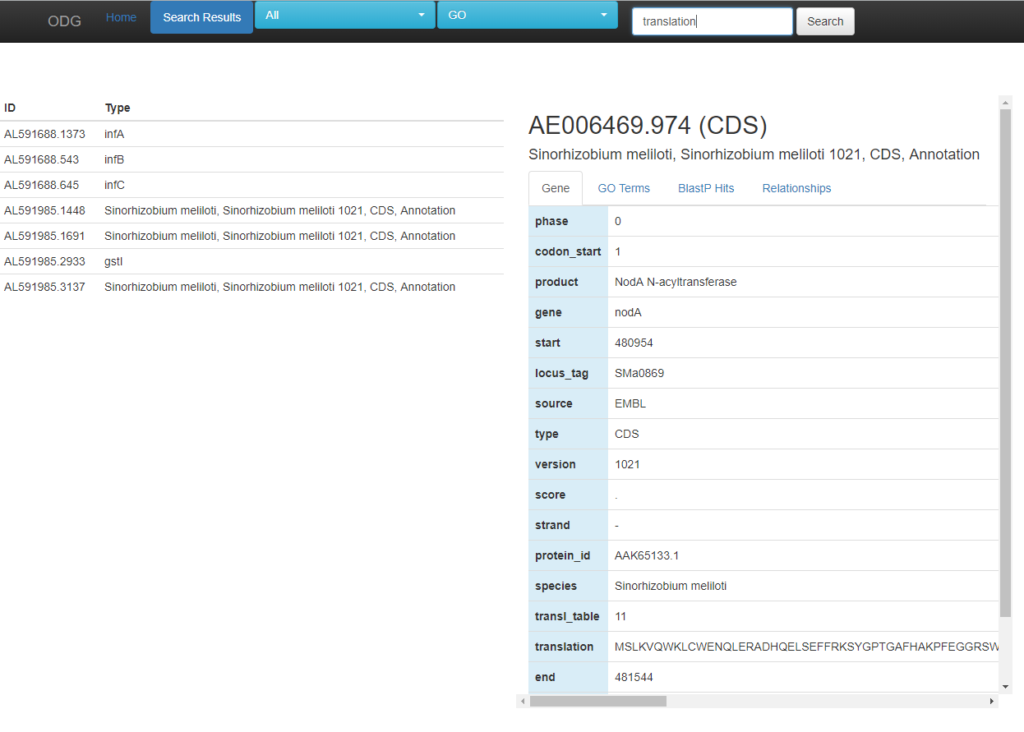
 After a search is completed, the results are listed on the left-hand side. Clicking on a gene (or other type of node) will bring up the appropriate page on the right-hand side. From here you can explore GO terms associated with the node, BlastP hits to other species, as well as additional relationships. Expression information, when included, is also found on this page.
After a search is completed, the results are listed on the left-hand side. Clicking on a gene (or other type of node) will bring up the appropriate page on the right-hand side. From here you can explore GO terms associated with the node, BlastP hits to other species, as well as additional relationships. Expression information, when included, is also found on this page.
Underlying Data Structure
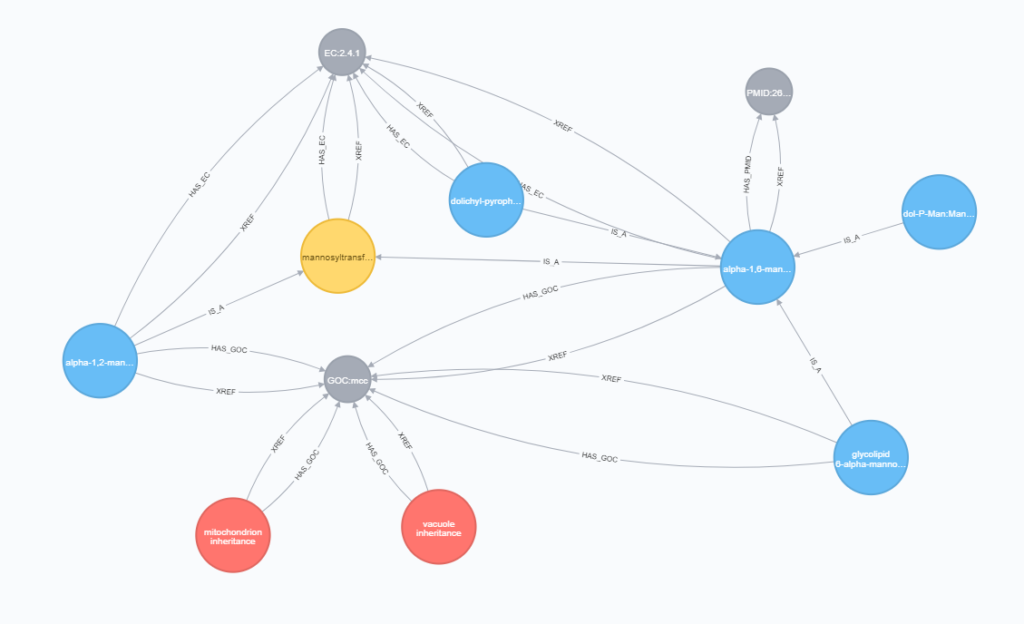
The strength of ODG comes from its graph-database backend, which can be accessed programmatically, without the ODG query interface. Here we can see an example of the rich data connections provided by the Gene Ontology database, which is integrated into ODG. Links from a species / accession can be made to GO terms through InterProScan analysis, PFAM analysis, or through a provided ontology association file. I have written a guide about connecting to the ODG generated database through the Neo4j Web Console.