The ODG query interface should suffice for many operations, and the command-line interface supports only certain analyses. If you have more advanced queries to run, you can interact with ODG’s generated database from nearly any programming language, using a library or package, via the REST API, or through Neo4j’s Web Console. This tutorial will cover accessing it via the Neo4j web console and is a pre-requisite for accessing it via programming languages.
Neo4j is the back-end graph-database server software that services ODG. ODG imports, exports, and analyzes data using Neo4j as the storage database. Thus, the ODG generated database works with Neo4j’s tools directly.
Mounting ODG to the Neo4j Web Console
To begin, please download the Test Drive version of ODG from the releases page. You may also use your database but will likely have to change some queries. You will also need to download the Neo4j server software from their website. ODG 1.1.0 uses Neo4j 3.2.1, although a newer version should work, it may take a few minutes to upgrade your database.
Make a copy of your database folder. We will mount ODG’s generated database using the Neo4j software directly. The database directory is specified in the configuration step and defaults to “odg.db”. For the Test Drive version, it is found in the extracted folder as “odg.db”. Make a copy of this folder somewhere, call it “odg.db.R” or “odg.db.copy” depending on your intended use.
If using windows, run the Neo4j server from the start menu and choose the directory where you have copied it.
If using Mac or Linux, move your copied folder to the Neo4j extracted directory and rename it to “graph.db” (removing any existing graph.db directory first). You may then start up neo4j using the command “./bin/neo4j start” from the command-line.
Running Queries via the Web Console
Point your web browser to http://localhost:7474/ once your databse is running, and login (username: neo4j password: neo4j then create a new password as the letter “a” for simplicity).
To try it out, run:
MATCH (x:Protein) RETURN DISTINCT(x.species)
That query will return a list of all species in the database with protein entries. The above command is the CYPHER query language, which we will use to interact with the database. You can read more about it at Neo4j’s website.
For a more advanced query try running:
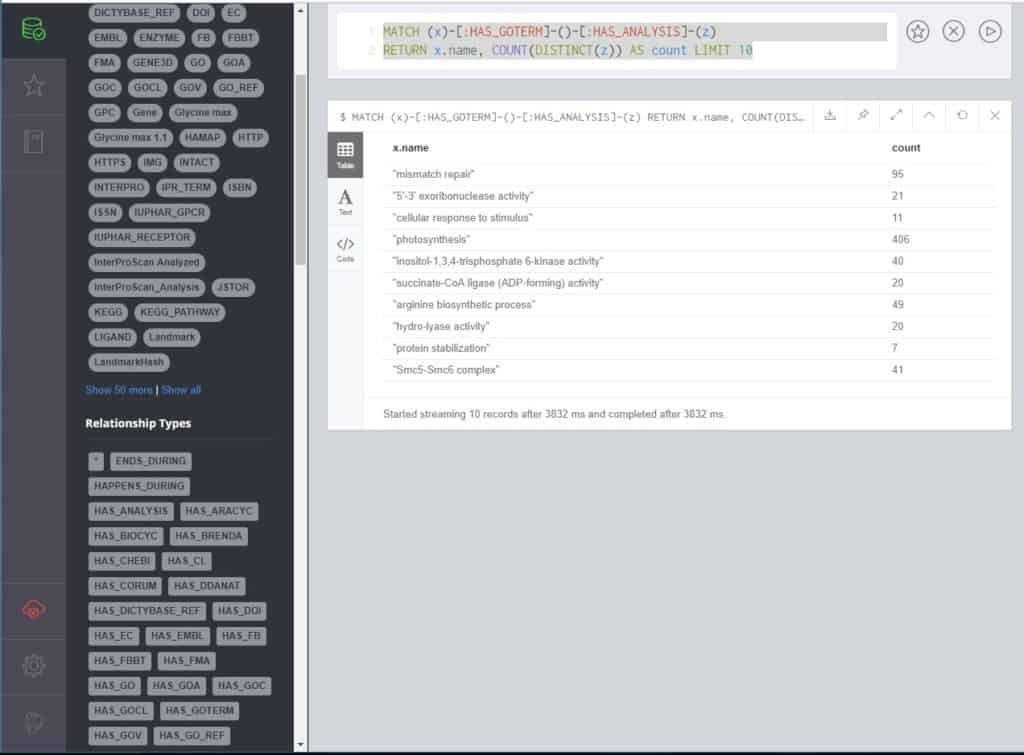
MATCH (x)-[:HAS_GOTERM]-()-[:HAS_ANALYSIS]-(z) RETURN x.name, COUNT(DISTINCT(z)) AS count LIMIT 10
This query returns 10 Gene Ontology (GO) terms and the count of genes in the database that correspond to that term. Because there are multiple paths to a GO Term (via InterProScan, usually, from different analyses), we only count DISTINCT entries. It’s important to note that we are only pulling in GO terms in the order they appear in the database. If you wanted to find the most common, for example, the query would become this:
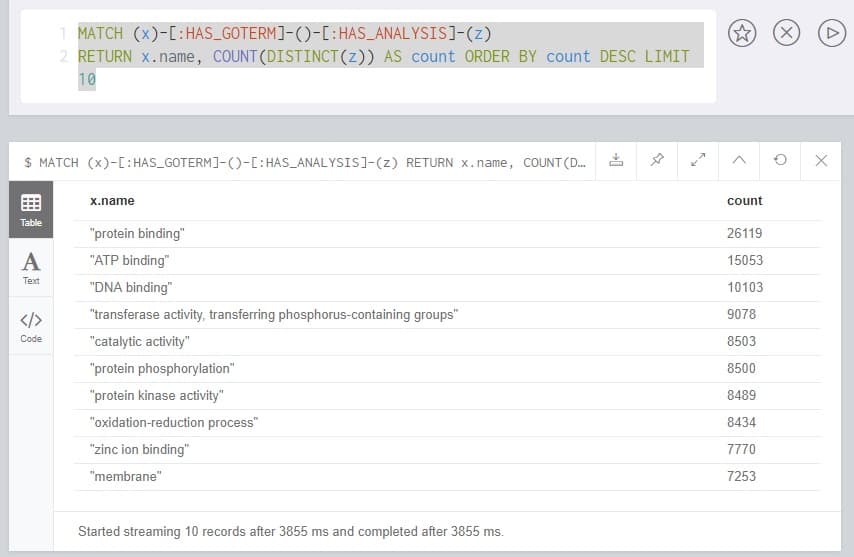
MATCH (x)-[:HAS_GOTERM]-()-[:HAS_ANALYSIS]-(z)
RETURN x.name,
COUNT(DISTINCT(z)) AS count
ORDER BY count DESC
LIMIT 10
This query should be fast but may take a little longer, as the database must calculate the entire problem space before ordering and returning the top ten entries.
Identifying Specific Genes
If you wanted a list of all genes annotated with the GO term for “mismatch repair” you could execute this query.
MATCH (x)-[:HAS_GOTERM]-()-[:HAS_ANALYSIS]-(z)
WHERE x.name = "mismatch repair"
RETURN DISTINCT z.id
If you wanted mismatch repair genes in Soybean (Glycine max) you could do this:
MATCH (x)-[:HAS_GOTERM]-()-[:HAS_ANALYSIS]-(z)
WHERE x.name = "mismatch repair"
AND z.species = "Glycine max"
RETURN DISTINCT z.id
If you wanted to identify Soybean genes with the PFAM entries “LRR domain binding” this is how the query would change:
MATCH (x)-[:HAS_PFAM]-()-[:HAS_ANALYSIS]-(z)
WHERE x.name = "LRR domain binding"
AND z.species = "Glycine max"
RETURN DISTINCT z.id
BLAST results are unfortunately absent from version 1.1.0 of the Test Drive due to GitHub space limitations, as the database becomes very large. Read up on the CYPHER query language to learn more. I will continue to add interesting queries to this tutorial as time goes on. Also, note there is a button to download a results table to a CSV, which can be imported into Excel, R, SAS, and many other languages and tools.
Pingback: This Week in Neo4j – 26 August 2017 – Cloud Data Architect