You can read this on medium.com, if you prefer.
Conda was a game-changer for bioinformatics, but it was slow. Then mamba came along and showed us speed. Now, Pixi is here to show you how fast things can really be. If you are a python developer, there are many reasons to use pixi. But here, I discuss it in the context of a bioinformatician. These are the reasons to use pixi, why to replace conda/venv with it, and how to get started.
For our purposes, Pixi is a direct replacement for conda/mamba. It handles creating and managing environments, installing packages, and cleaning up. It’s a small, self-contained binary; easy to install.
UV is a fast replacement for pip, allowing the installation of packages not on conda/bioconda. For most bioinformatics applications, you won’t need it, but for newer or more esoteric things, you may.
Speed, Isolation, HPC Compatability, and Reproducibility
Speed. Remember the early days of conda? Letting the solver run over lunch, or even overnight, just to install a package? While those days are mostly over in modern conda, there is still plenty of room for improvement, and Pixi brings it. You can even run the open-source benchmark. I’ve run it; check out below.
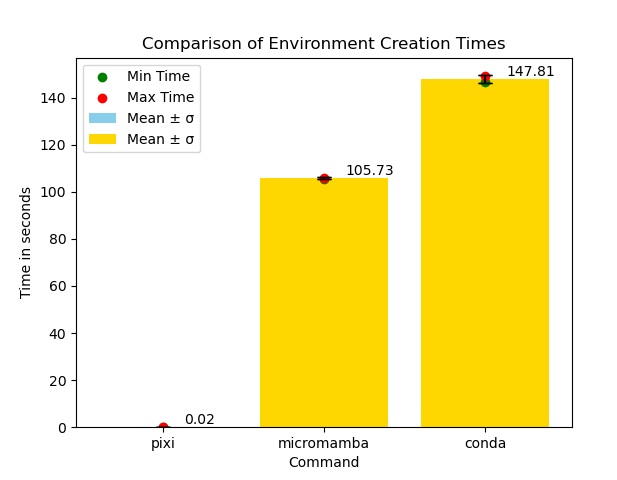
What a difference! It’d be unbelievable if I hadn’t been using it regularly.
Click to see the dummy_env.yml I used for this benchmark
channels:
- conda-forge
dependencies:
- scipy
- polars
- ipywidgets ==7.8.0
- seaborn
- matplotlib
- plotly
- polars
- hifiasm
- assembly-stats
- uv
- dgenies
- pip:
- pandocIsolation. One of the biggest benefits of conda are environments. But there is also a catchall shared “base” environment for ‘global’ tools. Pixi instead installs each global tool in isolated environments, so your base env is never destroyed. In fact, your environment is stored in the directory your environment works in, not your home directory. Which brings us to the next point…
HPC Compatibility. By default, conda puts all your environments in your home directory. However, most HPC clusters have a small quota for home directories, and these are often not backed up. Not to mention, as a diligent researcher, this creates an additional step of exporting your environment before committing it to GitHub or your other repository of choice. This additional separation not only makes the HPC admins happier but makes your projects far more portable and reproducible.
Reproducibility. By storing your pixi config and environment in your project directory, it makes commits a breeze, keeps a logical separation of environments, and makes it easy to wipe out the environment and rebuild it when needed. This is important if, say, a project is going down for a few months while in review, and then you need to adjust some parameters are rerun. If you download a project from github, or you’re going to an older project and have wiped the env to save space, you only have to run pixi install to install the environment.
Getting Started
Install pixi (or see the manual):
curl -fsSL https://pixi.sh/install.sh | bashSet default channels. Pixi comes with conda-forge by default, but if you here, you probably also want bioconda. The following command adds it to pixi’s default configuration; thus, all new environments will have bioconda and conda-forge as their default channels.
pixi config set default-channels '["conda-forge", "bioconda"]'Create an env. Let’s create a test environment.
# Run these commands
mkdir pixi_test
cd pixi_test
pixi initThis file is created:
❯ cat pixi.toml
[project]
authors = [“Joseph Guhlin email@address”]
channels = [“conda-forge”, “bioconda”]
description = “Add a short description here”
name = “pixi_test”
platforms = [“linux-64”]
version = “0.1.0”
[tasks]
[dependencies]
And just like that, we have our first environment.
Install some tools.
pixi add assembly-stats sra-tools ncbi-datasets-cliRunning a command. To run a command that is based in a pixi environment, prepend it with “pixi run” as below, which downloads an H5N1 genome and the following command to extract it. If you don’t have unzip, you can pixi add unzip and then run it with pixi run unzip ncbi_dataset.zip.
pixi run datasets download virus genome accession PP839258.1
unzip ncbi_dataset.zip
pixi run assembly-stats ncbi_dataset/data/genomic.fnaThat will get you the assembly-stats for this virus.
Conda activate equivalent. You can run things with pixi shell as well, which is more like running conda activate. Here, we download the next submitted H5N1 but without having to add pixi run to each command.
pixi shell
datasets download virus genome accession PP839259.1
unzip ncbi_dataset.zip
# Press A to overwrite all
assembly-stats ncbi_dataset/data/genomic.fna
exitExit at the end takes you out of the env’s shell.
Install a global tool. I use assembly-stats pretty much every day, and often without a specific project. So let’s install it as a global tool.
pixi global install assembly-statsThis installs it globally, so you can run it without pixi run. See more on global tools at the pixi docs here.
What about pip-only packages?
Pixi is to conda as UV is to pip. Again, we are talking speed. Pixi uses uv under the hood to install pypi packages now.
# Let's create a test env
mkdir pixi_test; cd pixi_test
pixi init
# Let's install python 3.11
pixi add python=3.11
pixi add --pypi jupyter jupyterlab numpy scipy polars pyarrowThat’s really it. Just like you would conda create -n myenv python=3.11 pip you can now do pixi init followed by pixi add python=3.11. A bit of verbosity for speed.
Pixi & uv will become even more integrated as time goes on, but there’s no reason not to use both now. You can even install uv as a global tool and create venv’s through that, and let pixi interact with them. I say keep it simple, come at this from a pixi-perspective, and realize uv is doing some work under the hood. But, if you need to:
Installing uv as a global tool
pixi global install uvPixi has extensive documentation, and guides for UV are available.