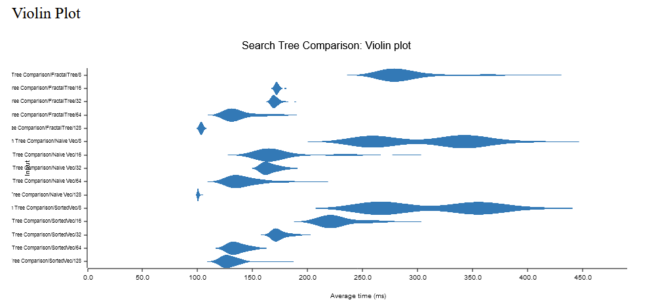
SFASTA: Fast Index building
What is SFASTA? Genomic and bioinformatic-adjacent sequences (RNA, Protein, Peptides) are stored as FASTA files. Sequencing reads off a machine are stored as FASTQ files, adding a quality score associated with each nucleotide. Currently, these are non-human-readable plaintext files. As sequencing increases, we need to be able to process many more gigabytes and terabytes of files rapidly and… Read More »